Figure 1
example.dat
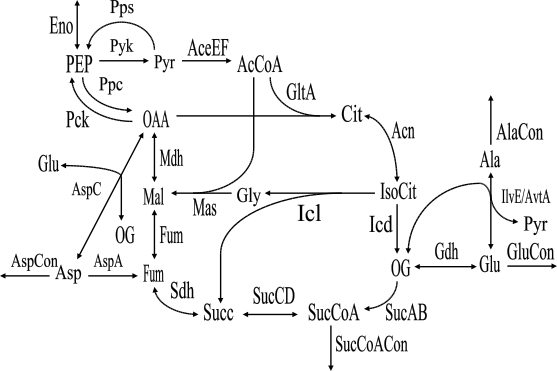
To explain the concept of elementary modes and the input file format of METATOOL, we give an example file (example.dat), which contains a reaction scheme comprising the tricarboxylic acid cycle, glyoxylate shunt and adjacent reactions of amino acid synthesis in E. coli (cf. Schuster et al. 1999). The network itself is shown in figure 1.

Figure 1
Two of the elementary modes in this network are shown in figure 2. They represent the usual TCA cycle (red) and an alternative catabolic pathway (blue), respectively. The latter has been predicted in Schuster et al. (1999) and recently been found in experiment by Fischer and Sauer (2003).

Figure 2
The different sections of the Metatool input file contain the following information:
-ENZREV, -ENZIRREV
After the key words -ENZREV and -ENZIRREV,
names or abbreviations of the reversible and irreversible enzymes,
respectively, have to be written.
-METINT, -METEXT
After the key word -METINT, names or
abbreviations of the internal metabolites have to be written. These
are the substances which have to fulfill a steady-state condition
(production = consumption). After the key word -METEXT names or
abbreviations of the external metabolites have to be written.
External metabolites (sources and sinks) need not be balanced in the
scheme under consideration. The order of these four fields is
important. All internal and external metabolites must have an
underscore or a letter (no number) as the first character and must
not include white space characters.
-CAT
After the key word -CAT, the reaction equations are
listed in any order. The raction name is written first in the same
way as after the key words -ENZREV and -ENZREV. The reaction name is
followed by a white space, a colon and a white space. Then the
stoichiometric reaction equation follows. Stoichiometric coefficients
are integers or floating point numbers separated by a white space
from the metabolites. After the metabolites, a white space and a plus
or an equal sign and a white space follow. The end of each reaction
equation is formed by a white space, a fullstop (optional in version
5) and a linefeed. Metabolites are written in the same way as after
the key words -METINT and -METEXT. The order of metabolites in the
reaction equations makes no difference. However, the sides of the
reaction equationsare exchangeable only in the reversible reactions.
The metabolites that are formed by the irreversible reactions have to
be written on the right side of the reaction equations.
The elements of the output file are described on this website.
The E. coli networks with biomass production
These are networks of the E. coli central metabolism including amino acid and fatty acid production which simulate bacterial growth on different media. This means that there is a growth reaction in each network which produces biomass from a list of precursor metabolites that are consumed in fixed proportions. These networks were first introduced by Stelling et al (2002) to examine the relation between structure and function in metabolic networks. In addition, they have been used for benchmarking purposes in several papers (e.g. Urbanczik and Wagner 2005, Klamt et al. 2005).
The Metatool input files that can be downloaded here were originally exported from the FluxAnalyzer and then modified to make the different external metabolites explicit. They differ in the types and combinations of substrates that are available as nutrients for the simulated metabolisms. The table below shows the number of elementary modes and computation times for these networks.
|
nutrients |
elementary modes |
computation time Metatool 5.0 |
|---|---|---|
|
acetate |
599 |
0.05 s |
|
succinate |
4250 |
0.3 s |
|
glycerol |
11333 |
1.2 s |
|
glucose |
27100 |
5.7 s |
|
combined: all 4 nutrients together |
507632 |
7 m 52 s |
References
E. Fischer, U. Sauer: "A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli", J. Biol. Chem. 2003 vol. 278 pages 46446-46451.
S. Klamt, J. Gagneur, A. von Kamp: "Algorithmic approaches for computing elementary modes in large biochemical reaction networks", IEE Proceedings Systems Biology 2005 vol. 152 pages 249-255
S. Schuster, T. Dandekar, D. A. Fell: "Detection of Elementary Flux Modes in Biochemical Networks: A Promising Tool for Pathway Analysis and Metabolic Engineering", Trends in Biotechnology 1999 vol. 17 pages 53-60
J. Stelling, S. Klamt, K. Bettenbrock, S. Schuster, E.D. Gilles: "Metabolic network structure determines key aspects of functionality and regulation", Nature 2002 vol. 420 pages 190-193
R. Urbanczik, C. Wagner: "An improved algorithm for stoichiometric network analysis: theory and applications", Bioinformatics 2005 vol. 21 pages 1203-1210